Fitness
Sodium chloride in the tumor microenvironment enhances T cell metabolic fitness and cytotoxicity – Nature Immunology
Cell purification and sorting
PBMCs from fresh peripheral blood of healthy donors were isolated by density gradient centrifugation using Ficoll-Paque Plus (GE Healthcare). Cytotoxic T cells and TH cells were isolated from the PBMCs by positive selection with human CD8-specific MicroBeads (Miltenyi Biotec), respectively, using an autoMACS Pro Separator (Miltenyi Biotec). Memory T cells were isolated as CD45RA− lymphocytes, and naive T cells were isolated as either CD45RA+CD45RO−CCR7+ lymphocytes or CD45RA+ T cells for scRNA-seq with a purity of >98%. The antibodies used for flow cytometry cell sorting have been described previously57,58,59. The cells were sorted with a BD FACSAria III (BD Biosciences), a BD FACSAria Fusion (BD Biosciences) or a Cytek Aurora CS.
Cell culture
Human T cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium supplemented with 1% (v:v) GlutaMax, 1% (v:v) nonessential amino acids, 1% (v:v) sodium pyruvate, penicillin (50 U ml−1), streptomycin (50 μg ml−1) (all from Invitrogen) and 10% (v:v) fetal calf serum (FCS; Sigma-Aldrich). For antigen-specific assays, FCS was replaced with 5% human serum (Sigma-Aldrich). Hypersalinity (high NaCl) was induced by increasing the NaCl concentration to 185 mM as described previously, unless indicated otherwise4,25. The final sodium concentrations in the cell culture medium were confirmed by potentiometry with a Cobas 8000 analyzer (Roche). T cells were stimulated with plate-bound CD3 (1 μg ml−1, clone TR66, Enzo Life Sciences) and CD28 mAbs (1 μg ml−1, clone CD28.2, BD Biosciences). T cell clones were generated under nonpolarizing conditions, as described previously, after single-cell deposition via flow cytometry-assisted cell sorting or by limiting dilution plating59,60.
Generation and isolation of antigen-specific T cells
To obtain tumor antigen-specific T cells (MART-1 specificity), 1 × 107 freshly isolated PBMCs were washed twice in phosphate-buffered saline (PBS) and resuspended in pre-equilibrated nucleofection buffer 1SM61. Then 20 μg of transposon-coding MART-1-specific TCR/0.5 µg of transposase SB100X solution was added to the cell suspension, which was subsequently transferred into the electrophoresis chamber. Nucleofection was performed on a Lonza Nucleofector IIb. After nucleofection, the cell suspension was transferred into a 24-well microplate containing complete culture medium with human serum and 50 U ml−1 of IL-2. After 24 h, MART-1-specific T cells were isolated from nucleofected PBMCs with a BD FACSAria III cell sorter by selecting CD3-FITC and anti-mouse-TCR β-chain APC (antigen-presenting cell)-positive cells after exclusion of dead cells with propidium iodide (PI).
MART-1-specific CD8+ T cells from the natural T cell repertoire were isolated from PBMCs from an HLA-A2-seropositive healthy donor after tetramer staining in MACS buffer (PBS supplemented with 1% (v:v) FCS, 2 mM EDTA) at 4 °C for 30 min. Tetramers were assembled the day before by incubating the peptide major histocompatibility complex (pMHC) with streptavidin–APC or streptavidin–BV421 overnight at 4 °C. The tetramers used for staining were pMHC–streptavidin–BV421 and pMHC–streptavidin–APC. The antibodies used for staining were CD3-FITC, CD8-PE (CD8-phycoerythrin) and CD19-ECD (CD19-extracellular domain). To exclude dead cells, PI was used. Single-cell sorting was carried out on a MoFlo Astrios Cell sorter (Becton Dickinson) after gating on single CD3+CD8+CD19−PI− pMHC–streptavidin–APC+/pMHC–streptavidin–BV421+ lymphocytes. The pMHC complexes loaded with MART-1 peptide were refolded as described previously9,62. Antigen-specific T cell clones were generated as described previously4,57.
Cytotoxicity assay
The viability of A375 melanoma cells (target cells) was determined in real time using an xCELLigence SP Real-Time Cell Analyzer (ACEA Biosciences), which allowed the quantitative and continuous monitoring of adherent A375 melanoma cells (target cells) through the measurement of electrical impedance every 15–30 min. For baseline measurements, 100 µl of A375 growth medium was added to the 96-well E-plate. A375 melanoma cells were seeded on to the E-plate at a density of 5 × 103 cells per 100 µl of growth medium. After 24 h, 100 µl of culture medium was replaced by 10−7 M MART-1 peptide in 100 µl of fresh medium. After 1 h of incubation with the peptide, MART-1-specific CD8+ T cells, which had been preactivated in high or low NaCl conditions for 5 d (or no T cells as a control), were added at a 1:1 ratio in 100 µl after removal of an equal amount of the growth medium. All conditions were set up in technical replicates. Cell indices were monitored every 15–30 min for another 24–48 h on the xCELLigence System at 37 °C with 5% CO2. Values are represented as cell indices (CIs), CIs normalized to the start of the coculture (nCIs) or as cell lysis calculated according to the following formula: \({\rm{Cell}}\,{\rm{lysis}}=\frac{{\rm{n}}{\rm{CI}}({\rm{A}}375\,{\rm{only}})-{\rm{n}}{\rm{CI}}({\rm{sample}})}{{\rm{n}}{\rm{CI}}({\rm{A}}375\,{\rm{only}})}\).
Flow cytometric analysis
For intracellular cytokine staining, human T cells were restimulated for 5 h with phorbol 12-myristate 13-acetate (PMA) and ionomycin with brefeldin A being added for the final 2.5 h of culture (all Sigma-Aldrich). The cells were fixed and permeabilized with Cytofix/Cytoperm (BD Biosciences for cytokines, eBioscience for transcription factors) according to the manufacturer’s instructions. The cells were stained with anti-cytokine antibodies (for antibody list, see Supplementary Table 2) and were analyzed with a BD LSRFortessa (BD Biosciences), a CytoFLEX (Beckman Coulter), a MACSQuant Analyzer (Miltenyi Biotec) or a Cytek Aurora Analyzer. Flow cytometry data were analyzed with FlowJo software (Tree Star) or Cytobank (Cytobank Inc.). Cytokines in culture supernatants were quantified by ELISA (R&D Systems) or Luminex (Thermo Fisher Scientific) according to standard protocols. High-dimensional cell surface phenotyping of sorted human memory CD8+ T cells from four individual healthy blood donors was performed with a newly established 33-color panel for surface markers (Supplementary Table 2) on a Cytek Aurora Analyzer. The cells were analyzed on day 5 after stimulation with CD3 and CD28 mAbs under high and low NaCl conditions.
For phospho-flow analysis, CD8+CD45RA− T cells were collected on day 5 after stimulation with CD3 and CD28 mAbs and rested in FCS-free medium at 37 °C for 30 min. After incubation with anti-CD3 (clone TR66, EnzoLifeScience) for 10 min on ice, cells were stained with live/dead zombie dye (BioLegend). The cells were then washed and incubated with anti-mouse immunoglobulin (Ig)G F(ab′)2 fragment for the indicated time points. Immediately after F(ab′)2 incubation, cells were fixed in CytoFix buffer or Lyse/Fix buffer at 37 °C for 10 min, then permeabilized by pre-cooled BD Perm buffer (both from BD Biosciences) on ice for 30 min. The cells were then incubated with an Fc blocker (BioLegend) and stained for 30 min at room temperature. Samples were recorded on a CytoFLEX and analyzed with FlowJo (BD Biosciences).
For 2-NBDG uptake assay, cells were harvested and rested in glucose-free medium for 10 min at 37 °C before incubation in glucose-free medium containing 100 μM 2-NBDG (Invitrogen) for 2 h at 37 °C. After a live/dead staining, samples were recorded on a CytoFLEX and analyzed with FlowJo.
For the BODIPY FL C16 uptake assay, cells were harvested and washed with 20 μM fatty acid-free bovine serum albumin (BSA)–PBS and then incubated in fatty acid-free BSA–PBS containing 1 μM BODIPY FL C16 (Invitrogen) for 30 min at 37 °C. After a live/dead staining, samples were recorded on a CytoFLEX and analyzed using FlowJo.
Ca2+ flux measurement
T cells were collected at day 5 after prestimulation with CD3 and CD28 mAbs under low and high NaCl conditions. A total of 5 × 106 cells were resuspended in 500 μl of cell culture medium with 5 μg ml−1 of Indo-1 AM or 3 μM Fura Red AM (both from Life Technologies) and incubated in the dark at 37 °C for 30 min. The cells were washed, then incubated in 10 μg ml−1 of anti-CD3 (clone TR66, EnzoLifeScience) in imaging buffer (PBS with 0.5 mM Ca2+, 0.5 mM Mg2+ and 1 g l−1 of glucose) for 10 min on ice. Cells were then washed and kept on ice with 7-AAD (BD Biosciences) or Höchest (Sigma-Aldrich) until measurement. For each Ca2+ flux measurement, cells were diluted with 37 °C pre-warmed imaging buffer and analyzed with a BD FACSAria Fusion Flow cytometer (BD Biosciences) or CytoFlex Flow cytometer (Beckmann Coulter). After acquisition of the baseline fluorescence level, TCR crosslinking was induced by addition of 1.3 μg ml−1 of AffiniPure F(ab′)2 Fragment Goat anti-Mouse IgG (Jackson ImmunoResearch). The calcium flux ratio was determined by the Indo-1 ratio between 400-nm (Ca2+ bound) and 475-nm (Ca2+ free) readings, or the Fura Red ratio between violet laser (Ca2+ bound) and green laser (Ca2+ free) readings. The calcium flux ratio was calculated using FlowJo.
Metabolic assays
The mitochondrial function (oxidative phosphorylation) and glycolytic rate of CD8+CD45RA− T cells, which were prestimulated for 5 d with CD3 and CD28 mAbs under high and low NaCl conditions, were assessed after washing using a Seahorse XFp Analyzer (Agilent Technologies). T cells were resuspended in Seahorse assay medium (pH adjusted to 7.40–7.45) on XF96 cell culture microplates at a density of 2.5 × 105 cells per well. Cells were centrifuged for 5 min at 400g to adhere and form a monolayer at the bottom of the plate. All experiments were performed with four technical replicates. The XFe96 extracellular flux assay kit (Agilent Technologies) was used according to the manufacturer’s protocol with the addition of oligomycin, CCCP (carbonyl cyanide m-chlorophenyl hydrazone), antimycin A and rotenone at concentrations of 2 × 10−6 M, 1.5 × 10−6 M, 2 × 10−6 M and 2 × 10−6 M, respectively. Evaluation and calculation of mitochondrial and glycolytic indices were done with the Wave software 2.2.0 (Agilent Technologies). Glut-1 expression by T cells was analyzed by flow cytometry using anti-hGlut1-PE (R&D, cat. no. FAB1418P) or anti-hGlut-1-FITC (R&D, cat. no. FAB1418F). Fatty acid uptake was analyzed by flow cytometry using BODIPY FL C16 (Invitrogen, 1 μM, excitation/emission 505/512 nm on FITC channel). For ATP quantification, the CellTiter-Glo Luminescent Cell Viability Assay (Promega, cat. no. G7570) was used. The luminescent signal was measured using CLARIOstar (BMG Labtech) and normalized to the absolute cell number. For quantification of the Na+/K+-ATPase activity, a colorimetric ATPase Assay Kit (Abcam) was used. After extraction of ATPase by a standard protocol, it was incubated with phosphate substrate in the presence or absence of 10 μM ouabain (Sigma-Aldrich) at 25 °C for 30 min. Results were recorded at 620 nm using a SPARK reader (TECAN).
Nontargeted metabolic profiling of CD8+ T cells after stimulation with CD3 and CD28 mAbs for 5 d in high and low NaCl conditions was performed by LC–tandem MS (LC–MS/MS) at Metabolon according to standard procedures. Raw data were extracted, peak identified and quality control (QC) processed using Metabolon’s hardware and software. After normalization to cell count, log(transformation) and imputation of missing values, with the minimum observed value for each compound, analysis of variance (ANOVA) contrasts were used to identify biochemicals that differed significantly between experimental groups. Data were imported to Graphpad Prism (v.7-9) for visualization.
Gene expression analysis
For messenger RNA-seq analyses (bulk transcriptome), CD8+ memory T cells were isolated ex vivo as described above and stimulated for 48 h with CD3 and CD28 mAbs for a total culture time of 5 d in the presence of low and high NaCl concentrations (Gene Expression Omnibus (GEO) accession no. GSE232365). Total RNA was extracted from cells lysed in TRI reagent (Sigma-Aldrich) according to the manufacturer’s protocol. RNA was quantified using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific) and its quality was verified by an Agilent 2100 Bioanalyzer according to the manufacturer’s guidelines.
Library preparation for RNA-seq was performed using the TruSeq Stranded Total RNA Sample Preparation Guide (Illumina). The barcoded libraries were sequenced on a NovaSeq 6000 platform (Illumina) by Novogene with paired-end, 150-bp reads (PE150). Approximately 2 Gb of sequencing reads were produced on average per sample. The reads were mapped to the reference transcriptome built from the human genome assembly hg38 (GRCh38) using STAR v.2.6.1a (ref. 63). Transcripts were quantified with salmon v.0.11.3 in the alignment-based mode64.
Significant DEGs were identified using the R package DESeq2 (ref. 65), using a false discovery rate (FDR)-corrected significance threshold of 0.05. Of 60,623 tested genes, we identified 26,837 with a nonzero mean expression. Of these, 1,956 were significantly up- and 1,926 significantly downregulated. Plots were produced with the R package ggplot2 (ref. 66). The R package clusterProfiler v.4.6.0 (ref. 23) was employed for overrepresentation analysis, as well as GSEA and visualization of GO terms within the ontology ‘Biological Process’ and KEGG pathways. The enrichment plots show the top 20 entries for the significantly upregulated, downregulated and dysregulated genes, respectively, which were ranked according to the shrunken fold-change values calculated by DESeq2, as previously described65. Barcode plots of enriched gene signatures were generated with clusterProfiler and the package fgsea (v.1.25.2) for computing a normalized enrichment score (NES) and statistics based on an adaptive multilevel split Monte Carlo scheme. Heatmaps were generated using the R package ComplexHeatmap v.2.14.0 (ref. 67). Overlay of expression data over KEGG pathways was done with the R package pathview v.1.38.0 (ref. 68). Real-time quantitative PCR (RT–qPCR) analysis was performed using Taqman probes targeting NFAT5 and 18S with a CFX Real-Time PCR instrument (BioRad).
Transcriptomic profiling of human cancers from TCGA
Gene expression data of patients with cancer were obtained from TCGA. Nondiseased, tissue-specific expression data were collected from the GTEx project. Gene expression data from both projects were re-preprocessed by the same RNA-seq pipeline from the University of California, Santa Cruz (UCSC) RNA-seq Compendium69. The resulting gene expression data were made publicly available as a TCGA–TARGET–GTEx cohort (https://xenabrowser.net/datapages/?cohort=TCGA%20TARGET%20GTEx), which was used for subsequent transcriptomic profiling of human cancers. Data from the TARGET cohort (pediatric data: https://www.cancer.gov/ccg/research/genome-sequencing/target) was discarded. We downloaded transcripts per kilobase million (TPM)-normalized RNA-seq-based expression data for 60,498 genes from the Xena platform as well as associated metadata (https://toil.xenahubs.net/download/TcgaTargetGtex_rsem_gene_tpm.gz, https://toil.xenahubs.net/download/TcgaTargetGTEX_phenotype.txt.gz, downloaded 6 March 2023)1. By using data from the toil hub, it was ensured that data were recomputed with one pipeline. After filtering for tumor entities that included healthy control tissues, 8,972 ‘primary tumor’ samples and 727 ‘solid tissue normal’ samples of 9,019 patients (TCGA dataset) and 4,472 ‘normal tissue’ samples of 535 donors (GTEx dataset) across 25 tumor types were available. Expression data for healthy samples (‘solid tissue normal’, ‘normal tissue’) were pooled per tumor site. To enable GSEA, log2(fold-changes) of gene expression data were calculated per gene and tumor sites between mean TPM-expression data of tumor and healthy samples. Subsequent GSEA using all 1,956 upregulated genes of the salt signature and the log2(fold-changes)-ranked gene list per tumor site as input was performed using the R fgsea package to compute NES and one-tailed test-based P values for positive enrichment of the gene signature. Default parameters were used for calculating GSEA with the function fgsea::fgsea except for minSize (=10) and maxSize (=4000) and scoreType (=‘pos’).
For Kaplan–Meier test statistics, TCGA-based expression data of NFAT5 (normalized to TPM) was filtered from the Xena platform69. The R packages survival (v.3.5-5) and survminer (v.0.4.9) were used to compute and visualize Kaplan–Meier survival statistics and significance with the Peto–Peto algorithm, respectively. TCGA data for breast and pancreas cancer were filtered for the attribute sample_type = ‘primary tumor’; 81 samples (from 81 patients) were obtained for pancreatic adenocarcinoma and 113 samples (from 113 patients) for breast cancer with information on time (attribute ‘new_tumor_event_dx_days_to’) and expression of the gene of interest. Each TCGA cancer patient dataset was subsequently subdivided into high and low gene expression groups by using the function surv_cutpoint of the survminer R package with default values.
Single-cell mRNA-seq analysis
For scRNA-seq analysis of matched tumoral and peritumoral CD8+ T cells from patients with breast cancer, tissue samples from three patients with breast cancer (BC01, BC02 and BC03) were reanalyzed (GEO accession no. GSE114727)11. QC with removal of cells expressing + T cells were annotated with CellTypist v.1.3.0 using the pretrained model Immune_All_High.pkl. Cells assigned to a probability of being a T cell 2.5 and CD8B > 2 were identified as CD8+ T cells. Transcriptomic NaCl signatures were derived from either a bulk transcriptomic comparison of FACS-sorted human CD8+CD45RA− T cells stimulated with CD3 and CD28 mAbs for 5 d under high and low NaCl conditions (FDR +CD45RA− T cells stimulated with CD3 and CD28 mAbs for 3 d under high and low NaCl conditions (Padj + T cells from the scRNA-seq analysis of the three patients with breast cancer. For GSEA, genes were ranked according to their expression values using the rank_genes_groups() function provided by Scanpy and analyzed with gseapy v.1.0.2. Module scores were calculated with the Scanpy score_genes() function. For comparisons of tumoral versus peritumoral CD8+ T cells, statistical significance was determined using Wilcoxon’s rank-sum test with the alternative hypothesis = ‘greater’ for upregulated gene sets and ‘less’ for downregulated gene sets.
For scRNA-seq (GEO accession no. GSE232149), a library of human CD8+CD45RA−CD45RO+ cells and human CD8+CD45RA+CD45RO− cells from one donor that were stimulated with CD3 and CD28 mAbs for 3 d under high and low NaCl conditions was constructed with Chromium Next GEM Single Cell 5′ Reagents v.2 (Dual Index; 10x Genomics, Inc.). The library was sequenced on an Illumina NovaSeq 6000 Sequencing System (Flow Cell Type S4) according to the manufacturer’s instructions, with 150-bp, paired-end, dual-indexing sequencing (sequencing depth: 20,000 read pairs per cell). Read alignment and gene counting of the single-cell datasets were performed with CellRanger v.7.0.1 (10x Genomics, Inc.). For downstream analysis, the filtered barcode matrix (CellRanger multipipeline) was processed with the R package Seurat v.4.0.4. For QC, cells with unique feature counts >9,000 and a count value >80,000 were filtered out. The total counts were normalized to 10,000 reads per cell. Each gene was centered and scaled to unit variance. Doublets were removed using the R package DoubletFinder v.2.0.3. UMAPs were generated using the RunUMAP() function and Leiden clustering was performed using the FindNeighbors() and FindClusters() functions from Seurat. The top ten marker genes per Leiden cluster were determined with the wilcoxauc() function in the R package presto (v.1.0.0) and depicted in a heatmap using the R package ComplexHeatmap (v.2.12.1). Bray–Curtis dissimilarity was calculated using the vegan R package (v.2.6.4). Differential gene expression was evaluated using FindMarkers() and genes were declared as significant based on an adjusted P-value threshold of 0.05. GSEA for GO was conducted using clusterProfiler (v.4.4.4). Multiple testing correction was performed using the Benjamini–Hochberg method and significantly enriched GO terms were visualized in a dot plot with clusterProfiler (v.4.4.4).
In analyses represented by violin plots, the python package Scanpy v.1.9.1 was used as an alternative to the R package Seurat v.4.0.4. Doublets were predicted and removed using Scrublet v.0.2.3 as before. Cells expressing _variable_genes(). PCA was performed using the function pca() n = 30. Finally, the neighbors of each cell and UMAP were computed using the functions scanpy.pp.neighbors() and scanpy.pp.umap(), respectively. Module scores were computed using the function score_genes() provided in the python package Scanpy v.1.9.1.
For trajectory analyses, diffusion pseudotime was used70. The cell with the highest expression of the stemness marker gene TCF7 was chosen as the starting point for pseudotime inference. For validation, the analysis was repeated using the cell with the highest expression of the stemness gene set from a public dataset71. The function diffmap() and dpt() from Scanpy were used to compute the diffusion map representation and to assign pseudotime values to each cell in the dataset. For RNA velocity, the velocyto pipeline v.0.17.17 was used to obtain the pre-mature (unspliced) and mature (spliced) transcript information based on CellRanger output. The functions scv.pp.moments(), scv.tl.velocity() and scv.tl.velocity_graph() from scVelo v.0.2.5 were applied to recover RNA velocity and scv.pl.velocity_embedding_stream() was used for visualization. QC and preprocessing were performed using the python package Scanpy v.1.9.1. Genes with a minimum count of 1 were retained and doublets were removed using Scrublet v.0.2.3. Total count normalization was applied using normalize_total(). Data were log(transformed) using log1p() and highly variable genes were identified using highly_variable_genes().
ICP–OES
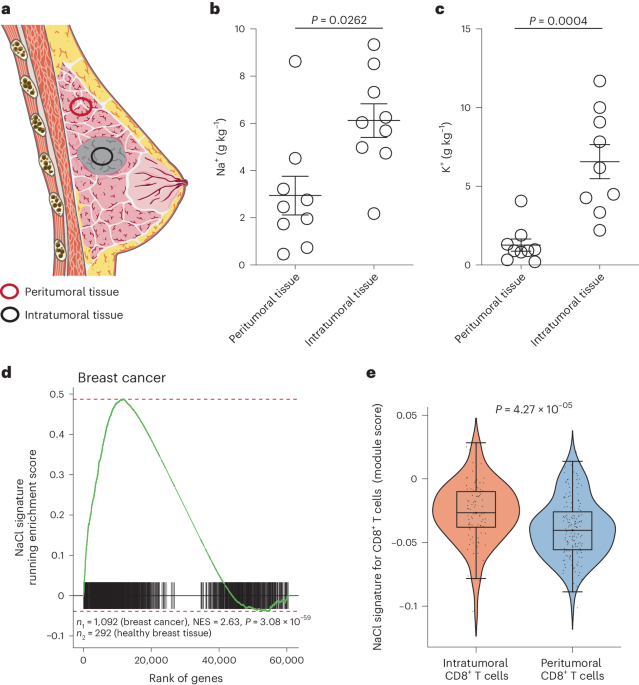
For sample preparation, frozen specimens from patients with breast cancer (age range 38–86 years) were subject to a freeze dryer (Heraeus Christ) and lyophilized at a −35 °C condenser temperature until the weight became constant. Subsequently, the dried samples were transferred into closed quartz vessels and digested with HNO3 (suprapure, sub-boiling distilled) in a Discover SP-D 80 microwave digestion system (CEM Corp.). The resulting solution was brought to exactly 10 ml with Milli-Q H2O and was then ready for element determination.
Element determination of Na and K was performed with ICP–OES ARCOS (Ametek-Spectro). The measured spectral element lines in nanometers were K 766.491 nm and Na 589.592 nm. Sample introduction was carried out by a peristaltic pump, connected to a MicroMist nebulizer with a cyclone spray chamber. The radiofrequency power was set to 1,400 W, the plasma gas was 15 l Ar per min and the nebulizer gas was 0.6 l Ar per min.
For QC, three blank determinations and a control determination of a certified standard (CPI) for all mentioned elements were performed regularly after ten measurements. Analysis of the results was carried out on a computerized laboratory data management system, which related the sample measurements to calibration curves, blank determinations and control standards.
Transmission electron microscopy
Cell pellets were fixed with Karnovsky fixative. After secondary fixation with 2% osmium tetroxide (Chemex) and 1% potassium hexacyanidoferrate (II) (Merck), serial dehydration steps were performed with acetone (Roth). Staining was performed with 1% uranyl acetate (Merck) in 50% acetone. Sample infiltration with Epon (Serva Electrophoresis) was performed with a mixture of acetone:Epon (3:1, 1:1, 1:3), followed by pure Epon and Epon with accelerator (BDMA, Agar Scientific). Samples were polymerized at 60 °C for 48 h. Trimming was performed with a Leica EM Rapid. Sections were made with an ultramicrotome UC7 (Leica). Semi-thin sections of 0.5 µm were dyed with Azure (Azure II, Sigma-Aldrich) and borax solution 5% (Sigma-Aldrich). Ultrathin sections of 55 nm were placed on copper slot grids coated with a Formvar/Carbon layer. The images were taken using a transmission electron microscope JEM 1400 (JEOL) with an acceleration voltage of 80 kV and the CCD camera ‘Orius SC 1000 A’ (GATAN) using the software GATAN Microscopy Suite (v.2.31.734.0). Images were analyzed using ImageJ2 (v.6).
Mouse experiments
Congenic C57BL/6 CD45.2−CD45.1+ mice and OT-1 (B6.Cg-Tg(TcraTcrb)1100Mjb) mice were bred in-house (Animal Facility Philipps-University Marburg, BMFZ). OT-1 (B6.Cg-Tg(TcraTcrb)1100Mjb) mice were obtained from Jackson Laboratories. CD8+ T cells were obtained from the lymph nodes and spleens of OT-1 mice using a negative selection kit (Miltenyi Biotec, cat. no. 130-104-075). For CTL differentiation, naive CD8+ T cells were cultured in RPMI (10% FCS) and stimulated with plate-bound mCD3 mAbs (3 µg ml−1, clone 145-2C11, BioLegend), soluble mCD28 (0.5 µg ml-1, clone 37.51, BioLegend), recombinant human IL-2 (50 U ml−1, Novartis) and anti-mIFN-γ (5 µg ml-1, clone XMG1.2, BioLegend) in the presence or absence of 30 mM NaCl. For intracellular cytokine staining, cells were restimulated after 72 h of culture with PMA (50 ng ml−1) and ionomycin (1 µg ml−1, both from Sigma-Aldrich) in the presence of brefeldin A (5 µg ml−1; BioLegend) for 4 h. Cells were fixed with 2% formaldehyde. Intracellular staining for TNF–FITC (eBioscience), granzyme B–PE (eBioscience) and IFN-γ–APC (BioLegend) was performed in saponin buffer (0.1% saponin and 1% BSA in PBS). For proliferation, CD8+ T cells were labeled with the cell proliferation dye eFluor 670 (2 µM). The cells were then washed, resuspended in culture medium and seeded into a 96-well-plate. Proliferation was measured by flow cytometry on day 3. Apoptosis was quantified using annexin V–PI staining. Cells were stained for 20 min in balanced salt solution with annexin V (APC, cat. no. 640920, BioLegend) at room temperature. PI (Invitrogen) was added immediately before flow cytometric analysis on an Attune NxT Cytometer (Thermo Fisher Scientific).
For in vivo animal experiments, 8- to 12-week-old CD45.1 mice were injected subcutaneously (s.c.) with 1 × 106 PancOVA cells. Then 7 d post-injection, tumor-bearing animals were treated with 0.5 × 106 CTLs, which had been differentiated from naive OT-1 CD8+ T cells for 3 d in vitro in the absence or presence of additional 30 mM NaCl. Tumor growth was measured and tumor volume was calculated (V = length × width2 × 0.5 mm3). For the ex vivo characterization of tumor-infiltrating T cells, tumors were isolated 3 d after adoptive T cell transfer and were digested with 200 U ml−1 of collagenase IV (Worthington, cat. no. LS004188), 10 μg ml-1 of DNase 1 (Roche, cat.no. 4536282001) in Hank’s balanced salt solution, 37 °C and 300 r.p.m. agitation and filtered through 100-μm cell strainers (Sysmex). Tumor lysates were stained with live/dead Zombie NIR dye (BioLegend, cat. no. 423106). Surface staining was performed using CD45.2 (BV510, BioLegend, cat. no. 109837), CD8 (PerCP/Cy5.5, BioLegend, cat. no. 100734), PD-1 (PE, eBioscience, cat. no. 12-9985-82) and TIM3 (APC, BioLegend, cat. no. 119706). For antigen-specific intracellular cytokine stimulation, tumor lysates were incubated for 4 h with OVA257–264 (JPT, cat. no. 43194) at 37 °C in RPMI with 5 µg ml−1 of brefeldin A. Intracellular staining was performed as described above.
Adherent PancOVA cells were grown in T75 flasks (Sarstedt) with Dulbecco’s modified Eagle’s medium (10% FCS) and split every 2–4 d on reaching 70% confluence, harvested by trypsinization (1× trypsin/EDTA, Sigma-Adrich, cat. no. T-4174) and reseeded (5 × 105 cells per 10 ml of medium in a T75 flask). For selection, 500 mg l-1 of G418 (Sigma-Aldrich, cat. no. G8168) was added.
CAR T cell generation
For CAR T cell manufacturing, CD8+ T cells were isolated with >90% purity from the spleens and lymph nodes of 8-week-old C57/BL6 mice by positive selection using magnetic microbeads (Miltenyi Biotec) and stimulated using plate-bound anti-CD3 (5 µg ml−1, clone 145-2C11) and soluble anti-CD28 mAbs (1 µg ml−1, clone 37.51) for 72 h. Then, 24 h after activation, CD8+ T cells were transduced with a second-generation ROR1 (tyrosine kinase-like orphan receptor 1) CAR using retroviral supernatant supplemented with polybrene, as described previously72. CAR T cells were expanded under high or low NaCl conditions for 48 h.
To assess the cytotoxicity of the anti-ROR1 CAR T cells, a coculture with firefly luciferase-expressing ROR1+ Panc02 target cells was performed. After washing with RPMI, CAR T cells and Panc02-ROR1 cells were plated at an E:T ratio of 10:1 and 2.5:1 (effector CAR T cell:target cell) in white, 96-well, flat-bottomed plates with medium containing 150 µg ml−1 of d-luciferin substrate. The chemiluminescence signal was measured at different time points at 37 °C using the Tecan plate reader Infinite 200. Lysis mediated by CAR T cells was calculated as the reduction of signal by effector cells compared with mock-transfected T cells: Specific lysis (%) = Mean((lysis by mock cells) − Single value (lysis by CAR T cells)/Mean(lysis by mock cells)) × 100. EpCAM-specific CAR T cells were generated using a second-generation CAR construct and functionally assessed according to a similar procedure.
Statistics
Statistical tests are indicated in the corresponding figure legends. Statistical tests for transcriptomic or metabolomic analyses are described in the corresponding sections. Error bars indicate the s.e.m. unless otherwise stated; P values ≤ 0.05 were considered significant; n indicates the number of biological replicates and individual blood donors. Analyses were performed using GraphPad Prism v.7-10. Data collection and analysis were not performed blind to the conditions of the experiments. For animal experiments, no statistical methods were used to predetermine sample sizes. Tumor-bearing mice were distributed equally between the groups according to their tumor size.
Study approval
Ethical approval was obtained from the institutional review boards of the Technical University of Munich (195/15s, 146/17s, 491/16s), Charité-Universitätsmedizin Berlin (EA1/221/11) and Friedrich Schiller University Jena (2020-1984_1). All blood donors provided their informed consent. All work was carried out in accordance with the Declaration of Helsinki for experiments involving humans and with the Regierungspräsidium Giessen for studies involving mice. The maximal tumor size of 1.5 cm, as approved by the Regierungspräsidium Giessen, was not exceeded.
Immunoblot analysis
Nuclear and cytosolic extracts from CD8+CD45RA− T cells after stimulation with CD3 and CD28 mAbs were separated using the Nuclear Extraction kit (Abcam). Protein concentrations were quantified using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Protein, 25 mg per sample, was boiled with 4× Laemmli sample buffer (BioRad Laboratories) containing 355 mM 2-mercaptoethanol (BioRad Laboratories) at 96 °C for 10 min and loaded on to a 8% sodium dodecylsulfate–polyacrylamide gel electrophoresis gel. The following antibodies were used: mouse anti-human NFAT5 antibody (Santa Cruz, cat. no. sc-398171), rabbit anti-human Lamin-B1 antibody (Cell Signaling Technology, cat. no. D4Q4Z), mouse anti-human β-actin (Cell Signaling Technology, cat. no. 8H10D10) antibody. Horseradish peroxidase-conjugated anti-mouse and anti-rabbit IgG antibodies (Cell Signaling Technology) were used as secondary antibodies. The immunoreactive bands were detected using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific). The chemiluminescence signals were recorded and analyzed using the iBright analysis system (Thermo Fisher Scientific).
Stable isotope tracing with metabolite extraction and GC–MS measurement
CD8+CD45RA− T cells were stimulated with plate-bound CD3 and CD28 mAbs for 48 h and cultured in complete medium until day 4 under high and low NaCl conditions as described above. The medium was replaced on day 4 with tracer medium containing 2 mM [U-13C-5]glutamine (Cambridge Isotope Laboratories) and all supplements were present in complete medium except for normal l-glutamine. Cells were then incubated for a further 24 h to reach isotopic steady state before starting metabolite extraction.
For metabolite extraction and the gas chromatography (GC)–MS measurement, the cells were washed with 0.9% NaCl. They were then quenched with ice-cold methanol and ddH2O (with 1 μg ml−1 of d-6-glutaric acid as an internal standard). Scraped cell extracts were combined with ice-cold chloroform, vortexed at 1,400 r.p.m. for 20 min at 4 °C, and then centrifuged at 17,000g for 5 min at 4 °C to achieve phase separation. Subsequently, 250 μl of the upper polar phase was transferred to GC glass vials with microinserts and subjected to cold vacuum drying. GC–MS measurement of isotope enrichment was performed. Dried extracts were derivatized at 55 °C with equal amounts of methoxylamine (20 mg ml−1 in pyridine) and MTBSTFA and injected into the GC–MS system. The separation of metabolites was accomplished using an Agilent 7890B gas chromatograph equipped with a 30 m × 25 mm × 0.25 µm ZB-35 ms (Phenomenex) and 5-m Guard column (Agilent). A sample volume of 1 µl was injected into the GC inlet at 270 °C using helium as the carrier gas at a flow rate of 1 ml min−1. The temperature of the GC oven was initially maintained at 100 °C for 2 min and then gradually increased by 10 °C per min until it reached 300 °C, where it was maintained for a further 4 min. The electron ionization energy was set to 70 eV. The temperatures of the MS source and the quadrupole were set to 230 °C and 150 °C, respectively. The detection of metabolites in selected ion mode was performed with an Agilent 5977 MSD system. The chromatogram analysis and the calculation of mass isotopomer distributions were performed with the Metabolite Detector Software73.
PPI analyses
To construct a PPI network for the mTOR pathway, curated PPI information was retrieved from BioGRID (v.4.4.230: https://thebiogrid.org) with the keyword ‘MTOR’ or ‘SGK1’. The resulting lists were put together and cleaned for duplicates. The R package igraph (v.1.4.2) was used to construct the interaction network. Node centrality was assessed by computing the node degree and visualized by adjusting node size. The log2(fold-change) of the bulk RNA-seq data (high salt versus low salt) was used to color the nodes. Edge colors were computed by Pearson’s correlation of CD8 bulk RNA-seq samples. Toward this aim, we computed first a log2(fold-change) per gene and RNA-seq replicate for high versus low NaCl, yielding three high NaCl-sensitive values per gene. Subsequently, the high versus low NaCl log2(fold-changes) were used to compute Pearson’s correlation for every gene–gene pair for which we found curated PPI information. In addition, STRING was used to construct functional PPI networks.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.







/cdn.vox-cdn.com/uploads/chorus_asset/file/25626295/247263_iphone_16_pro_AKrales_0799.jpg)
